Index .
Medical Science Knowledge Base
and (section 2) flow modeling
(3/19/2002)
Introduction
In this tutorial there are two downloads (1) and (2). The introductory tutorials and position papers are available.
Section 1: The proto-type work on a Medical Science Knowledge Base (MSKB).
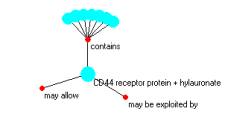
The data set consists of 813 triples of the form of a syntagmatic unit:
< a, r,
b>
The a and b be are often thought of as concepts and the r value as the relationship.
However, when syntagmatic units are assembled into a graph, we see that the role of concept and relationship can be flipped. The flip of roles is what Don and I call a conjugant map. These conjugants are seen clearly when navigating a complicated graph using the Event browser. In navigation, simple structures alternate between compounds (with a link in the middle) and atoms, with links not being in the middle.
One either places an atom in the middle or a link in the middle.
Part of the effect of the analytic conjecture is to produce the atoms and the links. Once these are produced, the warehouse has done its job. The “retail” products are the atoms.txt and links.txt files. These two files are all that is used by the SLIPCore browser. The atoms and links are encoded into a special memory map where some solid computer science allows the just in time composition of exactly those eventCompounds that are defined by the set of atoms and the set of links.
So the exercise here was to take the product of a specific 10-step process that Van Warren used to produce these 813 units. We move this structure into the process model for visual Abstraction (vA). We have the
1) Ability to visualize the categories that IS the data invariance
2) Ability to use the vA as a computational engine for various tasks
The trick to creating vA from the 813 triples is to break each triple into two pairs:
< a, r,
b> à (a, r), (b, r)
in doing so we create 1626 pairs. But, we find that there are only 371 unique relationships.
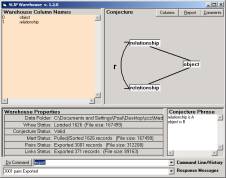
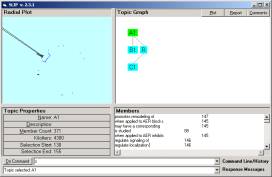
Figure 1: The analytic conjecture and SLIPCore
Our conjecture could have been (relationship, object) or (object, relationship); but we choose (relationship, object) so as to look at a question suggested by Van Warren regarding the categorization of the relationships.
We would like to be able to justify a minimal set of relationships through some sort of similarity analysis. But we put this issue in the back of our head, now, and just look at what we might find when we use only equality as a means to define categories. One can use a more advanced notion of similarity analysis using my work on declassification technology and the work on quasi-axiomatic theory.
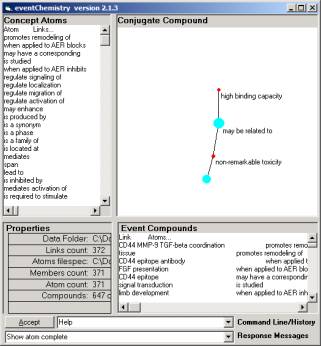
Figure 2: the Event browser
The current Event browser can only show either a simple compound (with the relationship in the center) or an atom.
Figure 3a is a compound with two relationships { high binding capacity, non-remarkable tonicity } and an unlabeled atom linked with non-remarkable toxicity.
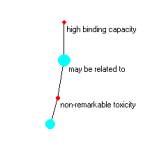

a b c
Figure 3: The Event Browser and three views of a compound
This construction corresponds to the syntagmatic unit:
< high binding capacity, may be related to, non-remarkable tonicity>
Remember, we flipped the object relationship designation using the Warehouse.
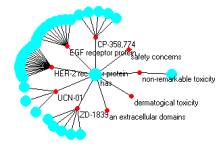
non-remarkable tonicity is related to the has atom. And the has atom is quite complicated as seen in Figure 3c.
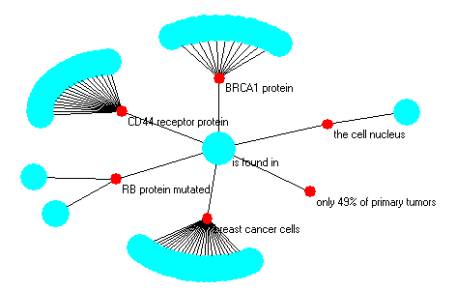
Figure 4: One of the relationship atoms
We face a number of problems. First is in regards to whether or not we might interface better with the set of syntagmatic units that are provided by Van Warren’s 10-step process. We will look into this as time permits.
There are a number of observations that we can make. As one example, look at Figure 3. The relationship is found in is a relationship that occurred in six of the pairs.
BRCA1 protein is found in
the cell nucleus is found in
RB protein mutated is found in
breast cancer cells is found in
CD44 receptor protein is found in
only 49% of primary tumors is found in
These pairs are produced from
< BRCA1 protein, is found in, cell nucleus >
< RB protein mutated, is found in, breast cancer cells >
< CD44 receptor protein, is found in, only 49% of primary tumors >
So the compound in Figure 3 cannot be read, for example as
< RB protein mutated, is found in, only 49% of primary tumors >
since this is not one of the facts that is in the original set of 813 triples. To treat this as a fact would be incorrect. It maybe, however, a conjecture and some automated reasoning system might extend the set of three facts, three syntagmatic units, to four facts. As a matter of a future discussion, one can extend the suggestions about conjecture, automated reasoning, and plausible reasoning.
So what do we have? We have visualized the concept of is found in, but we have to be careful in how we read the compound. But is this a matter of understanding how to use vA? I think that it is, but we have some more work to do. But this work moves us away from the AI Dream.
A personal comment can be made at this point. The attempt to make sense of vA in the context of this set of syntagmatic units could lead to a better understanding of what one might gain in the application of vA to other domains, such as:
- Cyber Defense Knowledge Base of Event Types
- Intellectual Property mapping and modeling of IP evaluation
- Business-to-Business value chain mapping.
- Use and improvement of the various paradigms for “machine ontology”
- Medical ontology use
- Etc.
What we need NOW is some capital formation to preserve what we have been able to do up to now.
- We have the BCNGroup as a not for profit Corporation.
- We have the for profit Corporation called OntologyStream Inc
The rules of law and economic theory allow us to use both of these legal entities. The BCNGroup can receive membership fees and donations, and this money can be used to support the mission of the BCNGroup as specified in the Charter;
BCNGroup Membership and
Charter
Section 2: Flow abstraction
We now look at a data set with a flow property. This flow property begins to show how visual abstraction (vA) might be animated to express uni-directional event expression such as seen in metabolic pathways. A relationship to Petri nets is indicated, both in Cyber Defense and in Bio and Chem warfare analysis and in basic research on medical sciences.
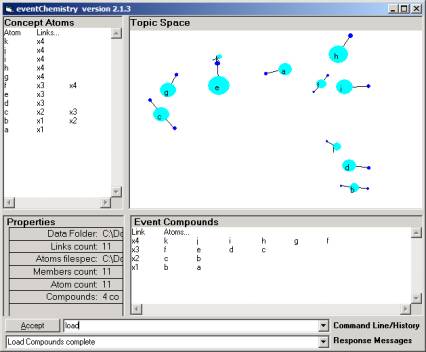
Figure 5: The eventChemistry browsers with 11 atoms
In Figure 5 we have 11 atoms scattered randomly into a three-dimensional object space. The rendering process, for these atoms, has been separated from the development of the object in memory structural holonomy. The object model is instantiated as an In-Memory Referential Information Base. The internal machinery of the I-RIB and a state-gesture Knowledge Operating System (root_KOS) is proprietary (trade secret) until June (2002), when the patent application will have been completed.
Our tutorial download contains only a datawh.txt file in the Data Folder. We will review how to use the SLIPWH (Warehouse) and SLIPCore. After downloading the 298 K zip file you may (in Windows) unzip to see Figure 6.
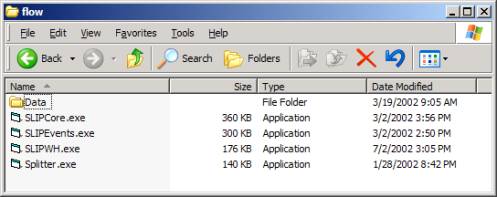
Figure 6: Flow.zip unzipped
Look into the Data folder to find only a small 1 K datawh.txt file. This file contains two columns. Using the Warehouse browser, you may issue the commands
· A = 0
· B = 1
· Pull
· Export
In order to produce Figure 7
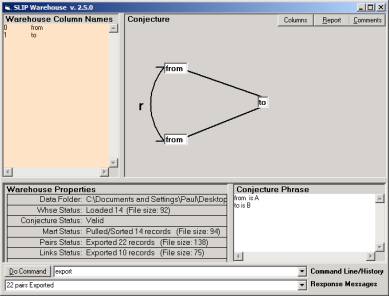
Figure 7: The Warehouse after commands issues via the KOS
As noted in previous tutorials, the KOS is a software component that can receive messages from various sources, and will respond to a message by parsing the grammar in the message. The source could be voice, for example.
Check in the Data folder and you will see 4 new files created. These files are to be used by SLIPCore.
Open SLIPCore and issue the commands
· Import
· Extract
Import produces the I-RIB and Extract produces the atoms.txt and maps this file into memory.
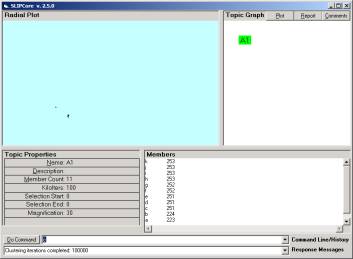
Figure 8: The SLIPCore after issuing commands via the KOS
Double clicking on the A1 node will take us immediately into the Event browser (see Figure 5).
The Event browser renders the I-RIB by responding to messages given to the KOS. Remember that the root-KOS is the foundational program for all SLIP browsers. We have separated the functional of the browsers mainly due to our need to exposit the underlying potentials of the architecture we have developed.
As verticals are developed for the OSI technology, then we will see various modifications to the browsers in order to restrict functionality and so to fit the SLIP technology into workflow. However the current browsers are tools that we use merely to explore the utility. We have explored this utility in tutorials and through the several proto-typing programs we have in place now (March 2002).
One should click on the links one at a time to see the relationships. One can image that animation will show the reaction change dynamically.
Various navigation ambiguities in a metabolic pathway is often due to response degeneracy (G. Edelman, “Neuronal Darwinism”) and these ambiguities depend on environmental conditions and “endophysics”. Thus the event chemistry can be used to reflect this endo and exo-physics. The foundations of much of the Soviet research in bio and chem. warfare used quasi axiomatic theory as a simulation theory and this work can be resurrected in the OSI Event detection systems.
As an exercise, one should copy the folder “flow” and delete all of the files and directories in the Data folder – except datawh.txt. Now open this file and cut and paste any part of the data records (except the first header line). Now repeat the commands that produce Figure 5. You will see that data is not visualized. It is the categories.
Because one adds no new categories to the atoms or links in repeating data, we have the clarity of looking only at the categories.
If something new occurs, then the formative refresh of the visual abstraction will immediate “see” the new categories of atoms and links. Retrieval is immediate, if this is required of data.
Therefore, this is a novelty detection system. I claim that this type of novelty detection is absolutely new and that this revolutionizes ALL military event detection and pattern recognition systems. It is simple, and thus almost not even patent-able.
What we have done that is patent-able is the rendering process using In-Memory Referential Information Base ™.